BrAPI R Tutorial
Overview
This tutorial will show you how to access data from T3 using BrAPI. The examples given here will be written in R, but the general ideas will be the same regardless of the programming language you choose to use.
What is BrAPI?
BrAPI is the Breeding Application Programming Interface - a standardized way for breeding databases and applications to transmit breeding data between themselves. It is a great way to programatically retrieve data from T3, other breedbase instances, and any other database that supports BrAPI.
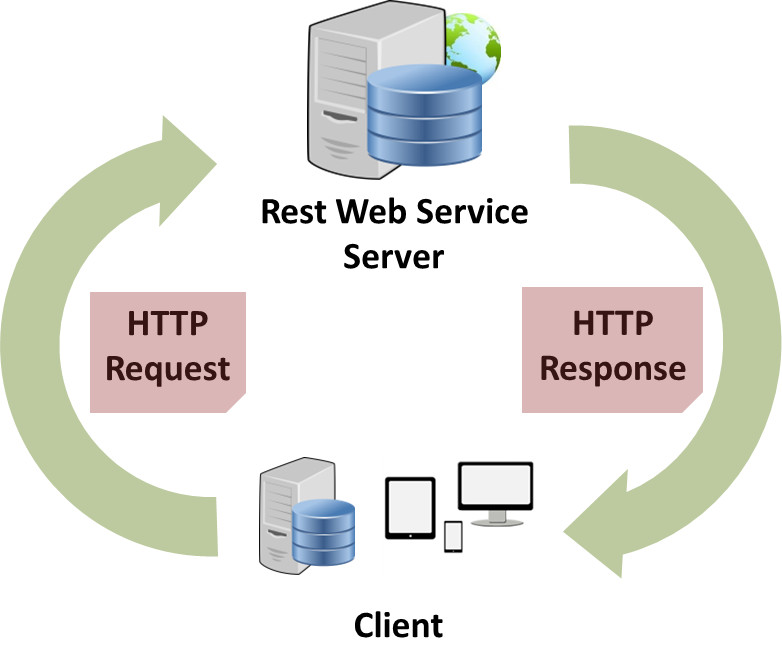
BrAPI is implemented as a RESTful API, meaning the client (the R scripts that you will write) makes standard HTTP requests
(using the httr R package) to documented BrAPI endpoints (the URLs available to access specific resources) on the server.
Ther server will then return a response that contains the data you requested, the status of a database search,
or information about the data you're adding to the database. The response is returned as text in a standardized
JSON format,
which R can parse into a named list for JSON objects and vectors for JSON arrays.
API Requests
A Request to an API server has the following components:
-
Method (required): the method describes the type of action your request will make on the server.
BrAPI uses these methods:- GET: when retrieving data from the database
- POST: when adding new data to the database
- PUT: when updating existing data in the database
-
URL / Endpoint (required): this is the URL pointing to the specific data resource you're requesting
Sometimes, the URL of the endpoint is dynamic and includes a parameter that is different depending on which specific resource you are requesting. For example, when requesting the data on a specific single germplasm record, the URL of the endpoint will include the unique germplasm ID. In the BrAPI documentation the endpoint may be represented as:/germplasm/{germplasmDbId}. In this example, the second part of the endpoint is a variable that you'll replace with a value when making your request, such as:/germplasm/2589. -
Query Parameters (optional): Additional key/value parameters that modify the request
For example, most BrAPI endpoints can acceptpageandpageSizequery parameters that determine which set of and how many result items are returned, for example:page=3andpageSize=25. -
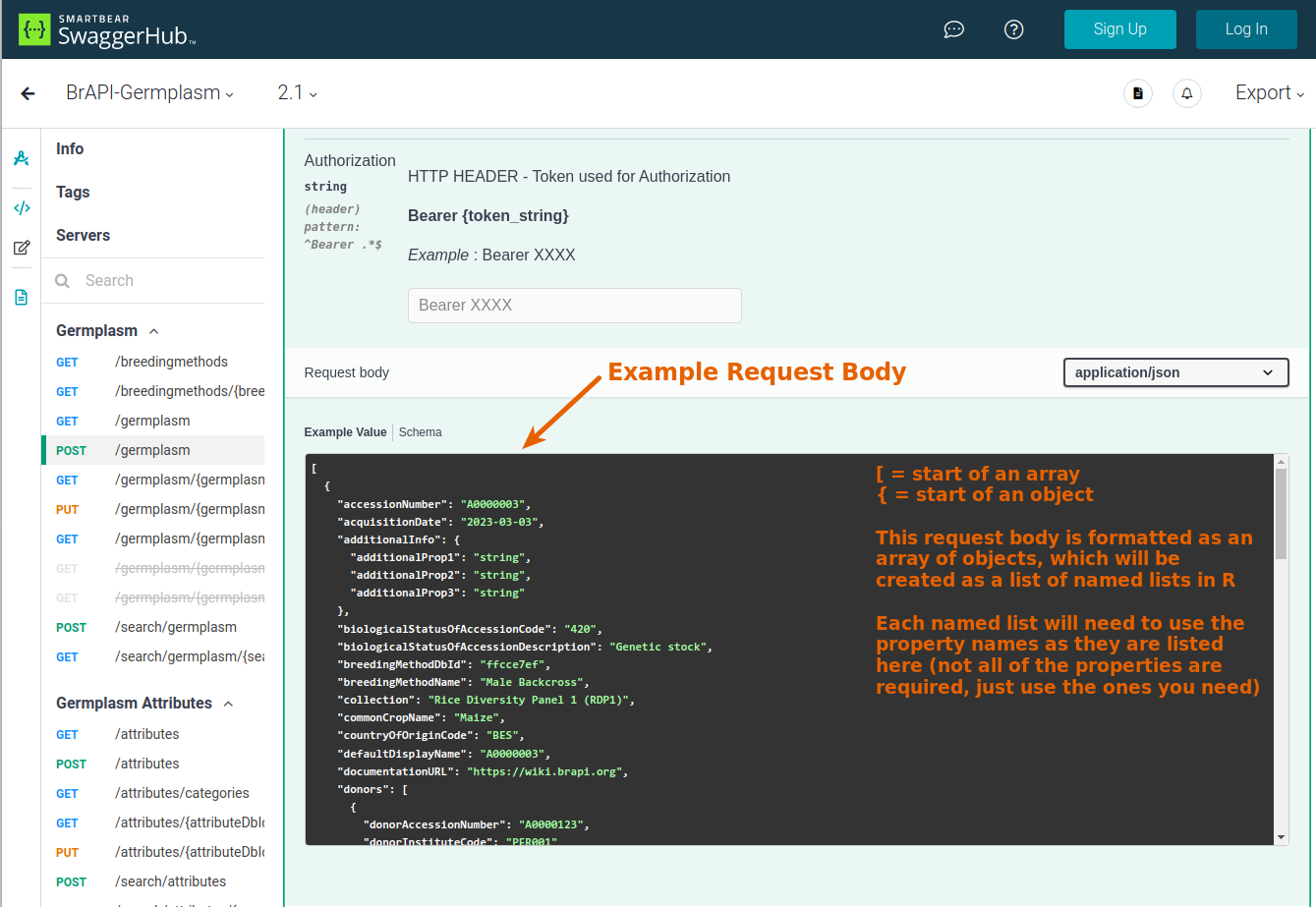
Body Parameters (optional):
POSTandPUTrequests generally include a body with the request, which is the content/data being sent along with the request (such as the new data being added to the database). In many RESTful APIs, including BrAPI, the the server expects the body to be formatted as JSON. In R, lists and vectors can be automatically converted into JSON before sending the request to the server.
BrAPI Endpoints
BrAPI has a large number of endpoints available to access all of the many different data types stored on T3. View the BrAPI Specification Documentation for a full list of BrAPI endpoints (some of which may not be implemented in T3/Breedbase).
The full URL of a BrAPI endpoint uses the following format:
https://server/brapi/version/call
- server is the host name of the specific database you're accessing, such as:
- wheat.triticeaetoolbox.org
- oat.triticeaetoolbox.org
- barley.triticeaetoolbox.org
- cassavabase.org
- sugarkelpbase.org
-
version is the version of the BrAPI specification you're using, such as:
- v1
- v2 -- which is the recommended version to use at this point
-
call is the final part of the URL / endpoint and specifies which resource you're requesting.
Some commonly used calls include:
- /germplasm
- /pedigree
- /studies
Usage
Install BrAPI R Package
Install the BrAPI.R package. This
package contains some simple boiler-plate code that handles GET, POST, and PUT
requests to a BrAPI server using the httr library.
# Install devtools, if you haven't already
install.packages("devtools")
# Install the BrAPI package from GitHub
library(devtools)
install_github("TriticeaeToolbox/BrAPI.R")
library(BrAPI)Setting the BrAPI Server
You'll need to create a BrAPIConnection object to send HTTP requests to a specific BrAPI Server:
# Use a known BrAPI Server
conn <- getBrAPIConnection("T3/Wheat")
# Manually set the BrAPI Server host
oat <- createBrAPIConnection("oat.triticeaetoolbox.org")Setting the BrAPI Version
By default, the BrAPIConnection will use version 2 of the BrAPI specification. If you want to change the version,
you can do so when creating a new connection
# You only need to do this if you don't want to use version 2 of the BrAPI specification
oat <- createBrAPIConnection("oat.triticeaetoolbox.org", version="v1")Finding a BrAPI Endpoint
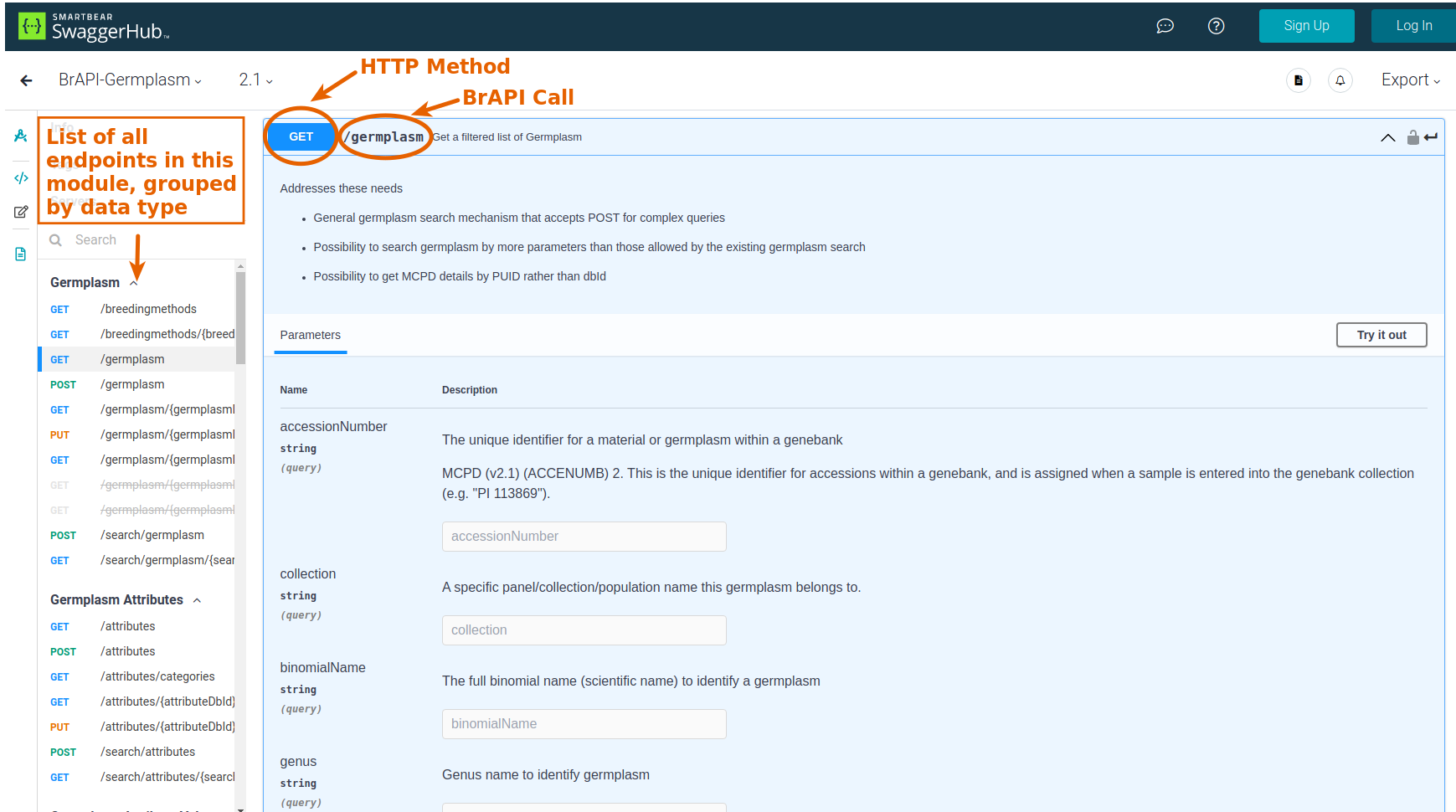
Use the BrAPI Specification Documentation to find a BrAPI Endpoint that will return the data you're interested in. The documentation is broken into different modules:
- BrAPI-Core: contains endpoints related to programs, trials, studies, and locations.
- BrAPI-Germplasm: contains endpoints related to germplasm, crosses, seedlots, pedigrees.
- BrAPI-Phenotyping: contains endpoints related to observations, observation units, observation variables (traits).
- BrAPI-Genotyping: contains endpoints related to calls, call sets, variants, variant sets, plates, samples, maps.
From the Documentation, you'll need to get:
- HTTP Method: this will be either
GET,PUT, orPOST -
BrAPI Call: this will be the final part of the URL to the BrAPI endpoint, such as
/germplasmor/search/trials
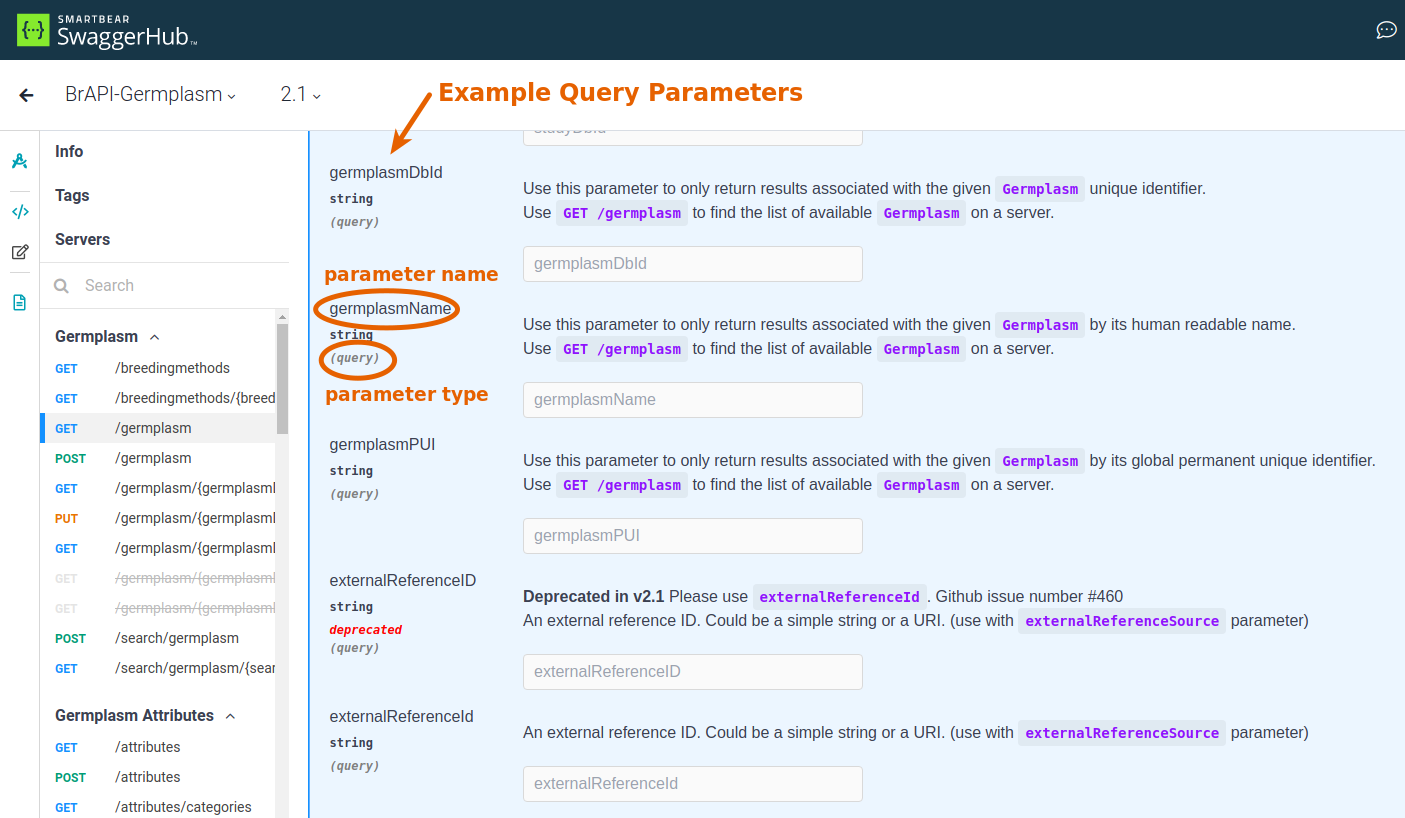
Each endpoint will have its own set of parameters that can be used to modify the request or send data to the server. To use a parameter, you'll need to know:
- parameter name: query parameters have a specific name that needs to be used and a request body needs to follow a specific format with properly named attributes.
- parameter type: this will either be listed as a query parameter or will be shown as part of an example request body.
Making a Request
Once you know the method, call, and any parameters, you can make a request to the specific BrAPI endpoint using one of the functions of the `BrAPIConnection` object. There is one function for each method:
- conn$get(call, ...): makes a GET request
- conn$put(call, ...): makes a PULL request
- conn$post(call, ...): makes a POST request
Each of the methods requires the call to be specified, such as /germplasm or /search/trials
Depending on the specific request you're making, you can add additional named arguments:
-
query: a named list of query parameters. For example:
query = list( programDbId = 360, species = "Triticum aestivum" ) -
body: a named list of the request body. For example:
# create a named list for the first item in the body (to be converted into a JSON object) b1 = list( observationUnitDbId = 1380641, observationVariableDbId = 84308, value = 125 ) # create a named list for the second item in the body (to be converted into a JSON object) b2 = list( observationUnitDbId = 1380563, observationVariableDbId = 84308, value = 130 ) # combine the two named lists into an unnamed list (to be converted into a JSON array) body = list(b1, b2) - page: specify the page number of paginated results (default: 0) or
allto fetch all available pages - pageSize: specify the number of items per page of paginated results (default: 10)
- token: specify the access token for requests that require authentication
Handling the Response
If the request was successful, the helper scripts will return a list with the Response properties
- response: the raw httr response
- status: information about the status code returned with the response
- content: the full data returned in the response.
- metadata: metadata about the response, such as pagination information and/or error messages (if returned in the content)
- data: the data of the response (if returned in the content)
If you requested all of the pages (page="all"), then all of the above response properties will be a list containing
the property for each returned page. It will also contain a vector named combined_data which will contain the data
from each response page combined into a single vector.
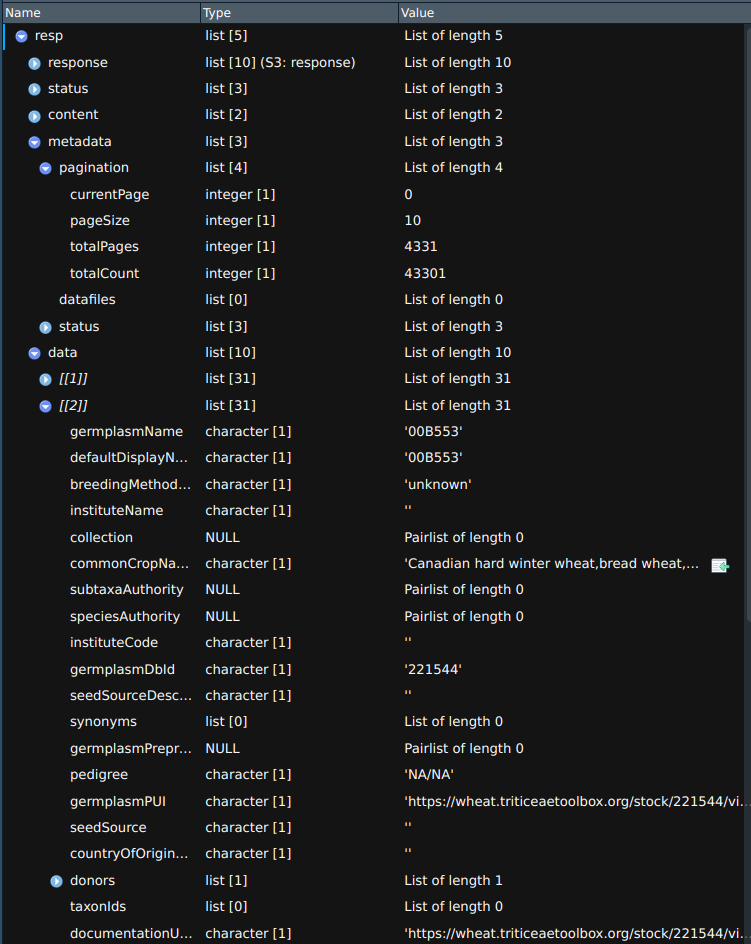
This screenshot shows the general structure of a BrAPI response. This response is from the /germplasm endpoint
and the r$data list contains information about the first 10 germplasm entries returned from the query.
The r$metadata$pagination list contains information about the current page and total number of pages.
Pagination
Many of the BrAPI requests can return any number of matching items from the database. To reduce the chances of getting
a very large response back, the results are paginated. This means that not all of the results will be returned at once.
By default, only 10 items will be returned per page. You can increase the number of items returned by setting a higher
page size in the request (by setting the pageSize argument). By default, the first page (page 0) is returned.
To get additional pages, you'll need to make additional requests and set the page argument in each request.
For example, to get 50 germplasm per page and to get the 2nd page (the first page is page = 0):
r = conn$get("germplasm", pageSize=50, page=1)When you set page="all", all of the available pages will be fetched and the properties of each page along with the combined
data will be returned from the request function.
r = conn$get("germplasm", pageSize=1000, page="all")
# a vector containing metadata for all 43000 germplasm
r$combined_dataAuthentication
Depending on the setup of the BrAPI server, some or all of the requests may require you to authenticate (login) to the
server before you can make any requests. Making a POST request that adds data to the database will generally
always require you to authenticate first. Retrieving data from the database may not require you to authenticate.
To authenticate with a breedbase server (the authentication process may differ for different BrAPI servers):
# send your username and password to the server to get an access token
l = conn$post("token", query=list(username="your_username", password="your_password"))
# extract the access token from the response
t = l$content$access_token
# t should be a long random alphanumeric stringThen, you'll use the access token in any future requests:
r = conn$post("observations", body=list(b1, b2), token=t)Searches
Many of the search endpoints, like /search/germplasm and /search/studies don't return
the search results right away. Instead, they return a searchResultsDbId which allows the search
to run in the background. Once you have the search ID, you make another request to /search/germplasm/{searchResultsDbId}, etc... to get the status of the search and its results once complete.
Examples
Germplasm
To get a single germplasm entry filtered by name:
r = conn$get("germplasm", query=list(germplasmName="JERRY"))
jerry = r$data[[1]]To get any matching germplasm entries by searching by name:
# start the search
s = conn$post("search/germplasm", body=list(germplasmNames=list("JERRY", "AJAX")))
# extract the search id from the first response
id = s$content$result$searchResultDbId
# get the search results
r = conn$get(paste0("search/germplasm/", id))
# extract the matching germplasm entries from the second response
matches = r$dataT3/Breedbase Trials = BrAPI Studies
To get all of the observations for a Study (you'll need to refer to the Study by its DB ID):
r = conn$get("observations", query=list(studyDbId=9411))The returned data will include one object for each recorded plot / trait observation for the specified Study.
For more examples, see the README.md or TUTORIAL.md files in the BrAPI.R repository.